TCR libraries for infection diseases and cancer
T cells evolved over millions of years to protect the host from infections through the specific recognition and efficient killing of diseased cells. This concept has been widely used therapeutically over the last decade to treat infections and tumors via the adoptive transfer of naturally occurring, antigen-specific T cells. Recently, the engineering of T cells with antigen-specific TCRs has opened up new possibilities for generating more reliable and versatile “living drugs”. This involves introducing a transgene encoding a TCR of interest into autologous patient-derived lymphocytes ex vivo. The resulting TCR-engineered T cells are then reinfused into the patient, where they can eventually recognize and eliminate target cells. Our goal is to contribute to the accessibility of TCR therapy on a broad scale by constructing libraries of virus- and tumor-specific therapeutic TCRs, alongside developing T-cell manufacturing processes suitable for clinical application.
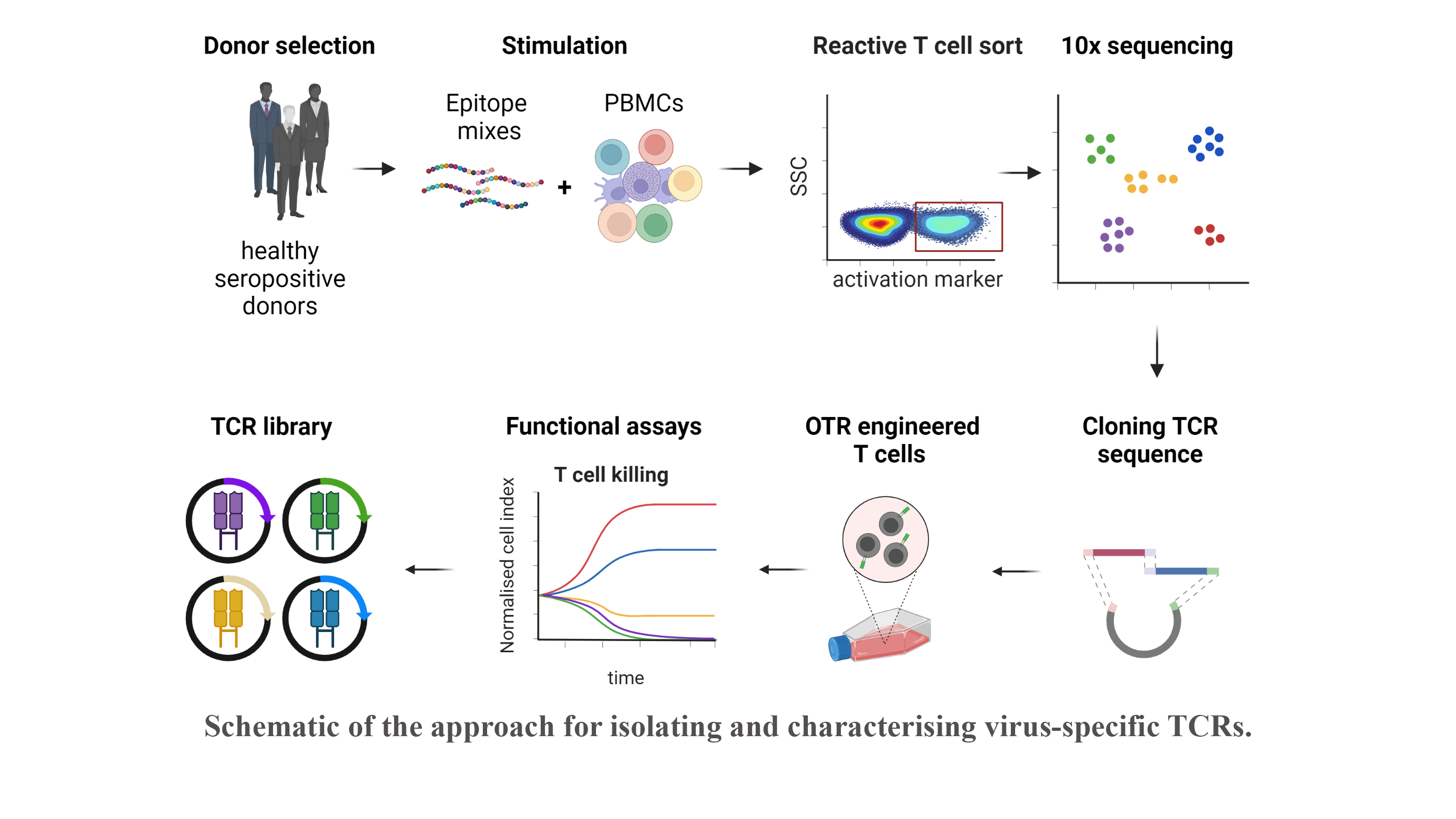
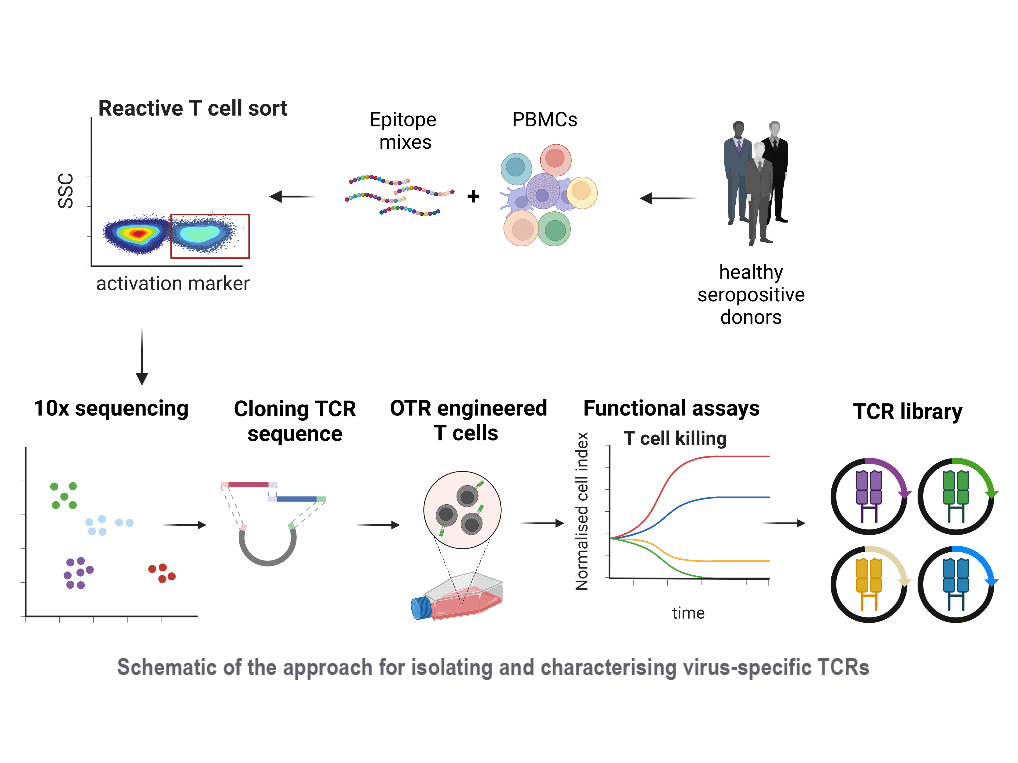
Reactivation of latent viral infections poses a significant threat to immunocompromised individuals, including those who have undergone hematopoietic stem cell transplantation. While antiviral medications are employed to manage these infections, the emergence of antiviral resistance remains problematic. Adoptive T-cell transfer represents a promising solution, specifically for viruses such as human cytomegalovirus (CMV), Epstein-Barr virus (EBV), and adenovirus (AdV). The transfer of virus-specific T cells from seropositive donors to immunocompromised patients has shown success in a large number of clinical studies worldwide, including those led by Prof. Busch at the Munich site. However, the isolation of these virus-specific T cells from suitable donors is still technically and logistically complex. A broad applicability can instead be achieved through the use of TCR-engineered T cells. Our current efforts involve identifying immunodominant viral epitopes and their corresponding human leukocyte antigens (HLAs) to render the vast majority of patients eligible for TCR therapy. Subsequently, we utilize these epitopes to stimulate T cells from donors and isolate responsive T cells based on activation markers. Corresponding TCRs are sequenced and subjected to downstream analysis and characterization to select potential candidates for therapy.
With this approach, our goal is to create an "off-the-shelf" TCR library, offering a versatile and accessible solution to effectively combat viral infections in immunocompromised patients.
Personnel

Related publications
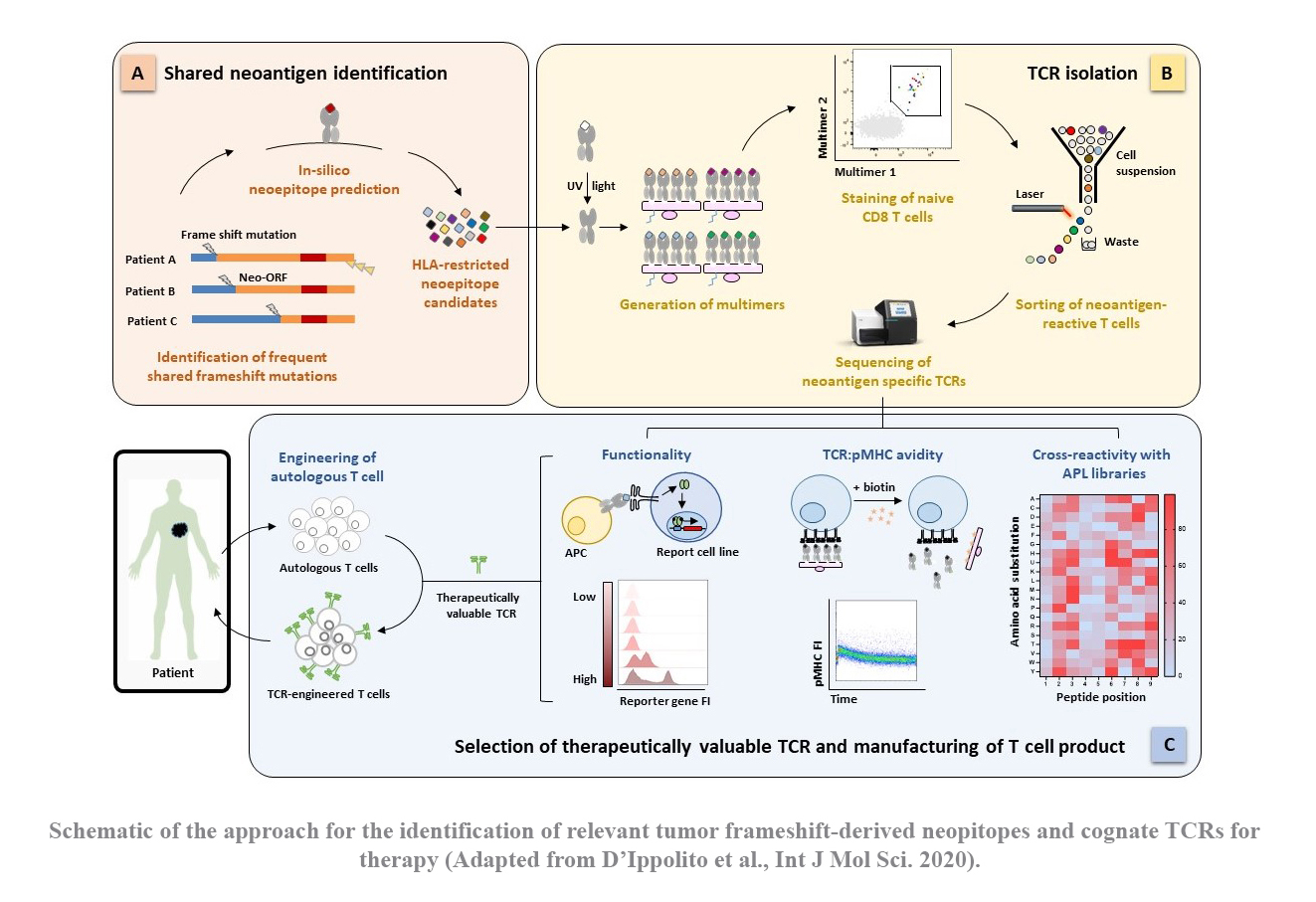
TCR-engineered T cells hold tremendous promise for adoptive cell therapy in cancer immunotherapy. Gastrointestinal cancers, such as pancreatic and colon cancers, present a significant challenge due to limited therapeutic options, driving the need for novel strategies. These tumors exhibit a deficiency in DNA mismatch repair mechanisms, resulting in an abundance of somatic mutations. These mutated proteins, known as ‘neoantigens’, are promising immunological targets due to their tumor specificity, thereby improving safety by minimizing off-target effects. Particularly intriguing are frameshift mutations caused by DNA insertions or deletions, which give raise to proteins with a large non-self-proportion. Moreover, the resulting neoepitopes could be potentially shared among different mutations and individuals.
In the past years, our group has successfully established a platform for identifying tumor-specific TCRs from naïve T cells of healthy donors (see section “How to find a therapeutic TCR”). Building upon this platform, our project aims to explore the landscape of frameshift-derived neoepitope in gastrointestinal cancers and subsequently identify functional TCRs for potential therapeutic use in clinical settings.
Personnel

Related publications
To make TCR therapy widely applicable, it is crucial to have a diverse array of TCRs that can recognize various viral or tumor epitopes in the context of different Human Leucocyte Antigens (HLAs). This is especially important in cancer treatment, where targeting multiple epitopes simultaneously can be advantageous in addressing tumor heterogeneity.
The advent of peptide major histocompatibility complex (pMHC) multimer technology has been revolutionary in T cell analysis, providing an unprecedented level of antigen specificity. Advancements in pMHC technology have further expanded the capacity for epitope discovery. Recently, barcoding of pMHC multimers with DNA oligonucleotides has enabled the simultaneous isolation of T cells with up to 1000 different epitope specificities. This multiplexed approach could significantly increase the pool of candidate TCR candidates for adoptive cell therapy that can be obtained from a single donor. A crucial step in this advancement was the development of UV-cleavable MHC ligands. With exchangeable UV-sensitive peptides, a single MHC sample can be loaded with one of thousands of different peptides to create a large pMHC multimer library in relatively short time.
In our T-cell isolation platform, we aim to leverage the power of cutting-edge pMHC technology, with a primary focus on identifying tumor-specific TCRs from the naïve T cell repertoire of healthy donors.
Personnel


